


China Surfactant Detergent & Cosmetics ›› 2021, Vol. 51 ›› Issue (4): 299-305.doi: 10.3969/j.issn.1001-1803.2021.04.006
• Development and application • Previous Articles Next Articles
GUO Yu-fei1( ),ZHANG Jian1(
),ZHANG Jian1( ),YU Wen2
),YU Wen2
Received:2020-06-25
Revised:2021-03-26
Online:2021-04-22
Published:2021-04-27
Contact:
Jian ZHANG
E-mail:18434390383@163.com;zhangjian@sxu.edu.cn
CLC Number:
GUO Yu-fei,ZHANG Jian,YU Wen. Molecular dynamics simulation of protease PB92 for washing before and after substrate binding[J].China Surfactant Detergent & Cosmetics, 2021, 51(4): 299-305.
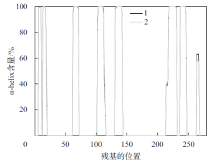
Fig. 4
Distribution of α-helix secondary structure before and after protease PB92 binding to substrate (1 indicates the distribution of α-helix secondary structure before protease PB92 binding substrate, 2 indicates the distribution of α-helix secondary structure after protease PB92 binding substrate)"
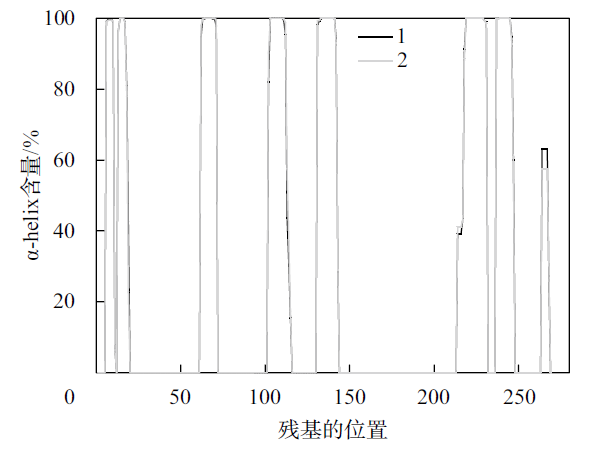
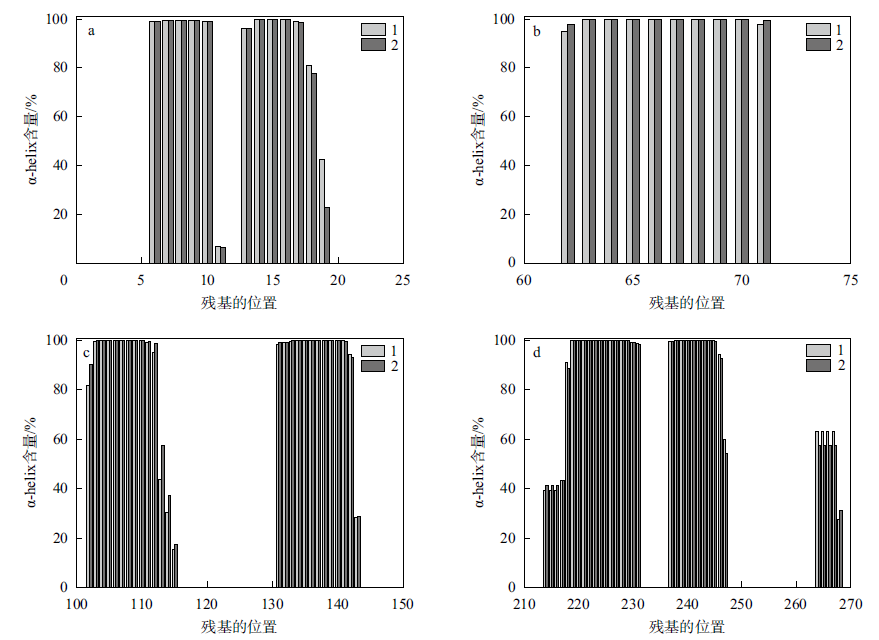
Fig. 5
Changes in α-helix content before and after the binding of protease PB92 to substrate (a. Amino acid No. 1 to 25; b. Amino acid No. 60 to 75; c. Amino acid No. 100 to 150; d. Amino acid No. 210 to 270; 1 indicates the change in α-helix content before protease PB92 binds to the substrate, 2 indicates the change in α-helix content after protease PB92 binds to the substrate)"
Tab. 1
Amino acids with significant changes in α-helix before and after protease PB92 substrate binding"
| α-helix含量/% | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 残基的位置 | |||||||||
| 19 | 102 | 113 | 114 | 247 | 264 | 265 | 266 | 267 | |
| PB92结合底物前 | 42.62 | 82.04 | 43.75 | 30.22 | 60.02 | 63.08 | 62.3 | 62.3 | 62.3 |
| PB92结合底物后 | 22.88 | 90.22 | 57.55 | 37.33 | 54.39 | 57.39 | 57.55 | 57.55 | 57.55 |
| 变化 | 19.74 | 8.18 | 13.80 | 7.11 | 5.63 | 5.69 | 5.68 | 5.68 | 5.68 |
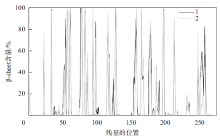
Fig. 6
The distribution of β-sheet secondary structure before and after protease PB92 binds to the substrate (1 represents the β-sheet secondary structure distribution before protease PB92 binds to the substrate, 2 represents the β-sheet secondary structure distribution after protease PB92 binds to the substrate)"
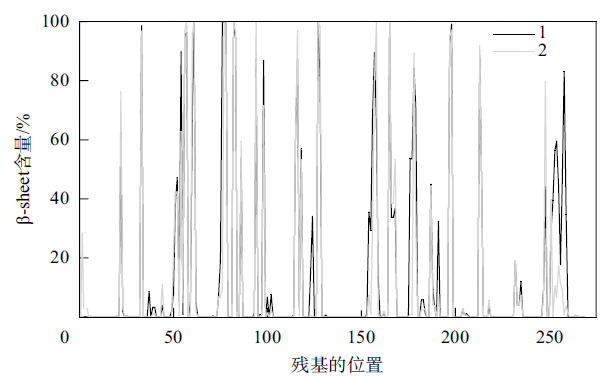
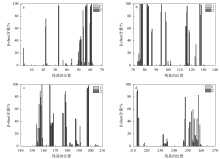
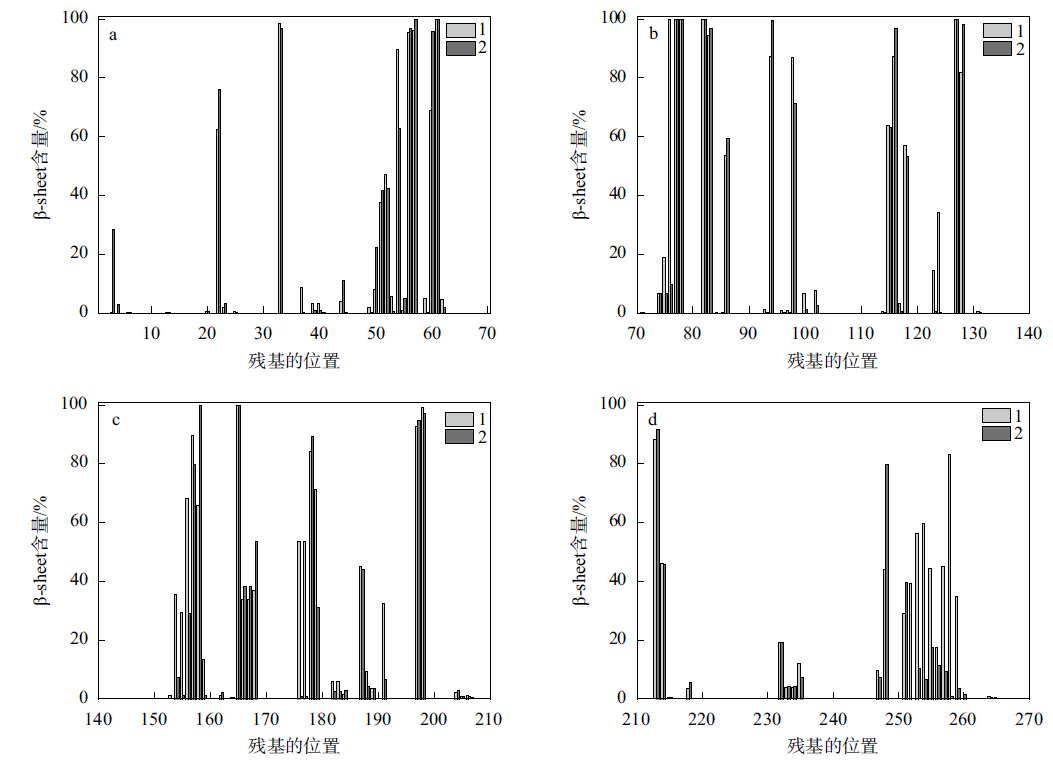
Fig. 7
The content of β-sheet before and after the binding of protease PB92 to the substrate (a. Amino acids No. 1 to 70; b. Amino acids No. 70 to 140; c. Amino acids No. 140 to 210; d. Amino acids No. 210 to 269; 1 represents the change in β-sheet content before protease PB92 binds to the substrate, 2 represents the change in β-sheet content after protease PB92 binds to the substrate)"
Tab. 2
Amino acids with significant changes in β-sheet before and after protease PB92 substrate binding"
| β-sheet含量/% | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 残基的位置 | |||||||||
| 158 | 176 | 179 | 248 | 252 | 253 | 254 | 257 | 258 | |
| PB92结合底物前 | 65.95 | 53.54 | 71.37 | 44.11 | 39.30 | 56.25 | 59.55 | 45.17 | 83.18 |
| PB92结合底物后 | 99.96 | 0.95 | 31.08 | 79.83 | 0.15 | 10.52 | 6.60 | 9.26 | 0.93 |
| 变化 | 34.01 | 52.59 | 40.29 | 35.72 | 39.15 | 45.73 | 52.95 | 35.91 | 82.25 |
| [1] | Tao Yan. Proteinase K molecular motion induced by substrate binding[D]. Kunming: Yunnan University, 2012. |
| [2] | Siezen R J, De V W M, Leunissen J A M, et al. Homology modelling and protein engineering strategy of subtilases, the family of subtilisin-like serine proteinases[J]. Protn. Eng., 1991,4(7) : 719-737. |
| [3] |
Blow D M. Structure and mechanism of chymotrypsin[J]. Accounts of Chemical Research, 1976,9(4) : 145-152.
doi: 10.1021/ar50100a004 |
| [4] |
Tsukada H, Blow D M. Structure of alpha-chymotrypsin refined at 1.68 Å resolution[J]. Journal of Molecular Biology, 1985,184(4) : 703-711.
pmid: 4046030 |
| [5] |
Neurath H. The versatility of proteolytic enzymes[J]. Journal of Cellular Biochemistry, 1986,32(1) : 35.
doi: 10.1002/jcb.240320105 pmid: 3533969 |
| [6] |
Martin J R, Mulder F A, Karimi-Nejad Y, et al. The solution structure of serine protease PB92 from bacillus alcalophilus presents a rigid fold with a flexible substrate-binding site[J]. Structure, 1997,5(4) : 521-532.
doi: 10.1016/s0969-2126(97)00208-6 pmid: 9115441 |
| [7] | Li Jing. Study on the interaction and application of metal calcium ion with alkaline protease[D]. Taiyuan: Shanxi University, 2018. |
| [8] |
Zhang Jian, Zhao Yongxiang. Study on the interaction between calcium ions and alkaline protease of bacillus[J]. International Journal of Biological Macromolecules, 2018.
doi: 10.1016/j.ijbiomac.2021.04.116 pmid: 33892028 |
| [9] | Yin Kai liang. Some basic applications and theories of molecular dynamics simulation[D]. Hangzhou: Zhejiang University, 2006. |
| [10] |
Yoon J, Park J, Jang S, et al. Conformational characteristics of unstructured peptides: alpha-synuclein[J]. Journal of Biomolecular Structure & Dynamics, 2008,25(5) : 505-515.
doi: 10.1080/07391102.2008.10507197 pmid: 18282005 |
| [11] |
Cordomí A, Ramon E, Garriga P, et al. Molecular dynamics simulations of rhodopsin point mutants at the cytoplasmic side of helices 3 and 6[J]. Journal of Biomolecular Structure & Dynamics, 2008,25(6) : 573-587.
doi: 10.1080/07391102.2008.10507204 pmid: 18399691 |
| [12] |
CalvinYu-ChianChen, Yuh-FungChen, Chieh-HsiWu, et al. What is the effective component in suanzaoren decoction for curing insomnia? Discovery by virtual screening and molecular dynamic simulation[J]. Journal of Biomolecular Structure & Dynamics, 2008,26(1) : 57-64.
doi: 10.1080/07391102.2008.10507223 pmid: 18533726 |
| [13] |
Zhao J H, Yang C T, Wu J W, et al. RING domains functioning as E3 ligases reveal distinct structural features: a molecular dynamics simulation study[J]. Journal of Biomolecular Structure & Dynamics, 2008,26(1) : 65-73.
doi: 10.1080/07391102.2008.10507224 pmid: 18533727 |
| [14] |
Macchion B N, Str?Mberg R, Nilsson L. Analysis of the stability and flexibility of RNA complexes containing bulge loops of different sizes[J]. Journal of Biomolecular Structure & Dynamics, 2008,26(2) : 163-173.
doi: 10.1080/07391102.2008.10507232 pmid: 18597538 |
| [15] |
Sonavane U B, Ramadugu S K, Joshi R R. Study of early events in the protein folding of villin headpiece using molecular dynamics simulation[J]. Journal of Biomolecular Structure & Dynamics, 2008,26(2) : 203-214.
doi: 10.1080/07391102.2008.10507236 pmid: 18597542 |
| [16] |
Mehrnejad F, Chaparzadeh N. Structural and dynamical studies of humanin in water and TFE/water mixture: a molecular dynamics simulation[J]. Journal of Biomolecular Structure & Dynamics, 2008,26(2) .
pmid: 18597538 |
| [17] |
Yoon J, Jang S, Lee K, et al. Simulation Studies on the stabilities of aggregates formed by fibril-forming segments of α-synuclein[J]. Journal of Biomolecular Structure & Dynamics, 2009,27(3) : 259-269.
doi: 10.1080/07391102.2009.10507314 pmid: 19795910 |
| [18] |
Cordomí A, Perez J J. Structural rearrangements of rhodopsin subunits in a dimer complex: a molecular dynamics simulation study[J]. Journal of Biomolecular Structure & Dynamics, 2009,27(2) : 127-147.
pmid: 19583439 |
| [19] |
Bairagya H R, Mukhopadhyay B P, Sekar K. An insight to the dynamics of conserved water molecular triad in IMPDH II (Human): recognition of cofactor and substrate to catalytic Arg 322[J]. Journal of Biomolecular Structure & Dynamics, 2009,27(2) : 149-158.
doi: 10.1080/07391102.2009.10507304 pmid: 19583440 |
| [20] |
Tai-Sung L, Cerutti D S, Dan M, et al. GPU-accelerated molecular dynamics and free energy methods in Amber18: performance enhancements and new features[J]. Journal of Chemical Information & Modeling, 2018.
doi: 10.1021/acs.jcim.1c00173 pmid: 33831302 |
| [21] | Pearlman D A, Case D A, Caldwell J W, et al. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules[J]. Computer Physics Communications, 1995,91:1-41. |
| [22] | Lee M C, Yong D. Distinguish protein decoys by using a scoring function based on a new AMBER force field, short molecular dynamics simulations, and the generalized born solvent model[J]. Proteins-Structure Function & Bioinformatics, 2010,55(3) : 620-634. |
| [23] |
Simmerling , Carlos , Hauser, et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB[J]. Journal of Chemical Theory and Computation: JCTC, 2015,11(8) : 3696-3713.
doi: 10.1021/acs.jctc.5b00255 pmid: 26574453 |
| [1] | LI Ju-long,DING Feng-mei,ZHOU Xiang,XING Zhi-qi,ZHAO Tao. Foamability of vanillin-based nonionic surfactants with different ethoxy chain length [J]. China Surfactant Detergent & Cosmetics, 2019, 49(7): 419-425. |
| [2] | CHEN Yu-jia, LIU Jia-xi, ZHU Qian-qian, HU Song-qing, SUN Shuang-qing, WANG Xiu-min. Effects of sodium salicylate on the self-assembly behavior of DTAB/PAM mixed system [J]. China Surfactant Detergent & Cosmetics, 2018, 48(6): 308-313. |
|